Neanderthal Man (16 page)

_______________________________________
While I was busy planning the new institute and Matthias Krings was trying to retrieve mtDNA from additional Neanderthals, the scientific community had begun to grapple with our analysis of the type specimen from Neander Valley. Our results did not go down well with proponents of the “multiregional-continuity” model of human origins who held, among other positions, that Neanderthals were among the ancestors of present-day Europeans. They shouldn’t have been so upset. In our 1997 paper, we’d carefully pointed out that while the Neanderthal mitochondrial DNA was clearly different from the mtDNA of any present-day humans, Neanderthals could nevertheless have contributed other genes—genes in the nuclear genome—to present-day Europeans. Perhaps the multiregionalists’ criticism of our work reflected a more general feeling of beleaguerment: as we were showing that, at least for the
mitochondrial
genome, the out-of-Africa and not the regional-continuity model applied, other researchers were finding that patterns of genetic variation in present-day humans supported the out-of-Africa scenario rather than the “multiregional-continuity” scenario. For example, our work was in good company, aligning with the work that Linda Vigilant, Mark Stoneking, and others in Allan Wilson’s lab had done in the 1980s on the mitochondrial genome. What’s more, we had begun to expand their work to the nuclear genome since I had moved to Germany. And the results seemed clear to me.
This work on the nuclear genome of present-day people was done by Henrik Kaessmann, one of the most talented graduate students I’ve ever known. Henrik came to the lab in 1997. He was tall, blond, athletic, and extremely serious about his work. I soon took a great liking to running with him in the Alps around Munich, especially up Hirschberg. (This mountain seemed to play a frequent role in my life.) After our strenuous runs up the winding logging roads and our leisurely jogs down them, we would spend time talking about science, especially genetic variation among humans.
We knew, from the work of Allan Wilson and others, that mitochondrial DNA variation was lower in humans than in the great apes, suggesting that humans were special in terms of having expanded from a small population. But we were acutely aware that the small size and simple inheritance of the mtDNA might be giving us a skewed view of the genetic history of humans and apes. By the time Henrik joined our lab, new and faster ways of sequencing DNA had made it possible to study parts of the nuclear genome in present-day people just as we and others had the mitochondrial genome. Henrik wanted to take on this challenge and study nuclear DNA variation in humans and in apes. But which part of the nuclear genome should he focus on?
We understand the function of only some 10 percent of the nuclear genome. These parts mostly contain genes that code for proteins. Such parts of the genome show very few differences between individuals because many mutations are harmful. Also, if a gene changed its function in the past so that the carriers of a new variant survived better or had more children, the gene may have spread in the population and show patterns of differences that reflect this. The rest of the genome is far less constrained by natural selection, presumably because these sequences do not have any essential functions that require DNA sequences to be preserved. Because we were interested in how random variation accumulates over evolutionary time, it was this 90 percent that was of interest to us. We chose to look at a particular region of 10,000 nucleotides on the X chromosome that contained no known genes or other important DNA sequences.
Having determined what part of the genome to sequence, we next turned to the question of which individuals to sequence. Males were the obvious choice because they have only one X chromosome (while females carry two), so Henrik’s task would be much simpler. But which males to sequence was a tougher choice. Others had often selected whichever people they had easy access to. For example, many genetic studies (generally of a medical nature) had been conducted using samples from people of European ancestry. A naïve user of databases of human genetic diversity might therefore believe that there is more genetic variation in Europeans than in other groups. But this, of course, may simply reflect the fact that groups other than Europeans had not been studied as much.
We could think of three ways to sample humanity more sensibly. First, we could collect our males based on how many people lived in different parts of the world. This, however, seemed a bad idea since our sample would be predominantly Chinese and Indians, large populations of whom resulted from developments during the past 10,000 years such as the invention of agriculture. In short, we would miss much of the world’s genetic diversity. Second, we could collect people according to land area, taking a sample every couple of square miles. But this, in addition to posing formidable logistic challenges, would result in over-sampling of sparsely populated areas such as the Arctic. The third option, which we finally adopted, was to focus on major language groups. We argued that major language groups (such as Indo-European, Finno-Ugric, and so on) reflect some approximation of cultural diversity going back more than 10,000 years. So by focusing on samples representative of major language groups, we could increase our chances of sampling most groups that have had long, independent histories. We would therefore hopefully cover more of human genetic variation.
Fortunately, others had come up with this idea before us so we were able to rely upon DNA samples collected by the distinguished Italian geneticist Luca Cavalli-Sforza at Stanford University. From those samples, Henrik selected sixty-nine men representing all major language groups and sequenced the 10,000 nucleotides in each of them. When he compared the DNA sequences in randomly chosen pairs of men, he found an average of just 3.7 nucleotide differences. Just as had been seen for the mtDNA, he found more variation between pairs of individuals from within Africa than from outside Africa. To gain some perspective on these results, he next turned to the closest living relatives of humans: the chimpanzees.
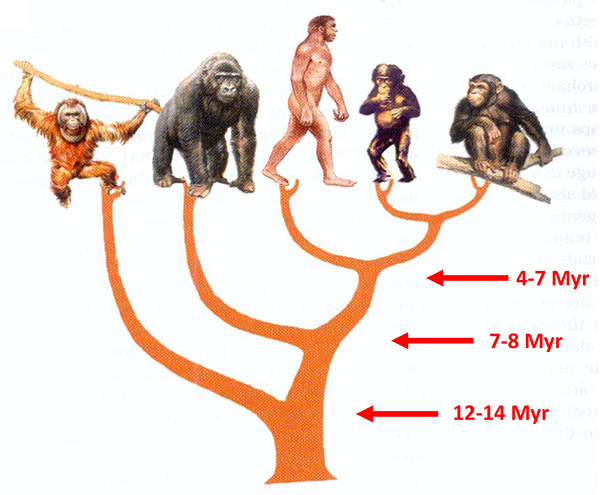
Figure 8.1. Tree of humans and great apes indicating approximate times when they may have shared common ancestors (although these dates are very uncertain). Modified from Henrik Kaessmann and Svante Pääbo “The genetical history of humans and the great apes,”
Journal of Internal Medicine
251: 1-18 (2002).
There are two species of chimpanzees, both living in Africa. The “common” chimpanzee lives in equatorial forests and savannahs in a patchy distribution stretching from Tanzania in the east to Guinea in the west, while the bonobo, sometimes called the “pygmy chimpanzee,” lives only south of the Congo River, in the Democratic Republic of Congo. Comparisons of DNA sequences had shown that the two chimpanzee species are the closest living relatives of humans, our lineages having split perhaps some 4 million to 7 million years ago. A bit further back, perhaps 7 million to 8 million years ago, humans and chimpanzees shared an ancestor with the other African great ape, the gorilla. Orangutans in Borneo and Sumatra share with the other great apes and humans an ancestor who lived perhaps 12 million to 14 million years ago (see Figure 8.1).
Henrik chose thirty male chimpanzees (the “common” species, not the bonobos), representing the major chimpanzee populations in eastern, central, and western Africa, and sequenced the same stretch of DNA on the X chromosome as he had in the humans. Again making comparisons between randomly chosen pairs, he found an average of 13.4 differences between any two individuals. It was, to my mind, an amazing observation. Seven billion humans hugely outnumber chimpanzees, perhaps numbering fewer than two hundred thousand. And humans live on almost every speck of land there is on the planet while chimpanzees live only in equatorial Africa. Yet any two chimpanzees carried three to four times as many genetic differences from each other than two random humans.
Henrik next sequenced the same piece of DNA in bonobos, gorillas, and orangutans to see whether humans are unusually similar to each other or chimpanzees unusually diverse. He found that gorillas and orangutans carry even more variation than the chimpanzees and that only bonobos had about as little variation as humans. We published these results in three papers in
Nature, Genetics,
and
Science
between 1999 to 2001,
{38}
demonstrating that a region in the nuclear genome had a pattern of variation very similar to what Allan Wilson’s group had found for mtDNA. The same pattern was likely typical of the entire human genome, and I became more convinced than ever that the out-of-Africa model for modern human origins was the correct one. So I listened to the critique of our Neanderthal work from the “multiregionalists” and was not impressed. But mostly I did not answer them. I was convinced that time would tell who was right.
Most multiregionalists were paleontologists and archaeologists. Although I did not dare say so publicly, I privately thought little about their ability to answer questions about whether one ancient group had replaced another one, mixed with it, or simply changed to become the other group. For the most part, paleontologists could not even agree on how to define the ancient groups they studied. There were—and still are—lively fights between “splitters,” who see many different species among hominin fossils, and “lumpers,” who see few. There are other problems inherent in paleontology. As famously stated by the anthropologist Vincent Sarich, who worked with Allan Wilson in the 1980s, we know that people living today had ancestors because they’re here, but when we see a fossil, we cannot know whether it had any descendants. In fact, most fossils we see in museums look like humans because they share ancestors with us sometime in the distant past, but they often have no direct descendants today and represent “dead-end” branches of our family tree. Yet there is often a tendency to think of them as “our ancestors.” In my enthusiastic moments, I imagined that the sequencing of DNA extracted from fossils would eventually do away with all this uncertainty.
One of our critics among the multiregionalists was the distinguished paleontologist Erik Trinkaus. He pointed out that our results would be biased if we erroneously discarded as contaminants any DNA sequences resembling those of present-day humans when found in Neanderthal bones. He argued that in fact these may be endogenous, true Neanderthal sequences. Certainly some Neanderthal bones had yielded
only
modern-looking sequences. But these were specimens with bad preservation, so I was confident that all endogenous Neanderthal DNA in them were gone and all we had seen were modern contaminants. Nevertheless, Trinkaus’s argument was a logical one and I felt we needed to address it directly.
This became the task of David Serre, a French graduate student from Grenoble with an enormous head of hair and a tendency to ski too fast down mountains in winter and canyon down powerful waterfalls in summer. We decided that his research, should he live to carry it out, would investigate whether all Neanderthals had mitochondrial DNA sequences similar to that from the type specimen, and whether early modern humans in Europe, who lived at the same time or slightly after the Neanderthals, lacked such DNA sequences. The latter question was important to pin down. As noted, the survival of a particular mtDNA sequence in a population is to a large extent influenced by chance. If early modern humans had arrived in Europe and mixed with the resident Neanderthals, then some, or even many, of them might have carried Neanderthal mtDNA sequences that could have become lost in subsequent generations if the females carrying them had no daughters. Indeed, soon after our 1997
Cell
paper appeared, Magnus Nordborg, a Swedish theoretical biologist working in the United States, had pointed out this scenario.
This criticism did annoy me, because it confused two separate questions. The first question was whether Neanderthals contributed mitochondrial DNA to modern humans that persists in people living today. We had answered this question in the negative. The second question was whether Neanderthals and modern humans interbred. This question we had not answered. However, I found the first question both more interesting and more important. I wanted to know whether I, or anyone else walking around today, carried DNA from Neanderthals in our bodies. If we had
not
inherited any DNA from the Neanderthals, any interbreeding 30,000 years ago was of no consequence from a genetic perspective. Whenever I spoke to journalists, I tried to make this point. To make it clear, I said that I was not in the least interested in sexual practices during the Late Pleistocene unless those practices had left any traces in our genes today. I sometimes added that I would be very surprised if modern humans had not had sex with the Neanderthals they encountered. But what mattered was whether they had children who lived to pass on their genes to us.
Despite my annoyance with these confused questions, I wanted David to investigate whether early modern humans in Europe might have carried Neanderthal mtDNA that subsequently became lost. If they ever did have such mtDNA, they would have carried nuclear DNA from Neanderthals as well. In that case, it would be reasonable to assume that some parts of Neanderthal nuclear DNA might linger on in people today.