Regenesis (31 page)
Authors: George M. Church

So Donnell and the three students, aided and abetted by Dr. Claudia Rocha, also of the Citadel's Biology Department, proceeded with handson microbial genetic tinkering. If and when they got the scheme to workâand even if they didn'tâthey would enter their project in the 2010 iGEM competition.
iGEM stands for international genetically engineered machines. The name had a stirring, thrilling, almost chilling ring to it. It was reminiscent of
R. U.R
., a science fiction play by the Czech author Karel Capek. It premiered in 1921, and gave the word “robot” to the world, for R.U.R. stood
for “Rossum's Universal Robots.” Capek's robots were not the metallic, tin can, electronics-driven devices of today. Instead, they were biological machines that had been assembled in a factory, part by part. In the play, unfortunately, the robots take over the world and wipe out the human raceâa tiresome and overworked scenario if there ever was one.
iGEM, by contrast, was a real-world organization in which student teams from around the world competed in building novel biological machines, also part by part. These “machines” were in fact microbes, but they were so substantially altered and enhanced by the genes of other organisms that they constituted new and original types of microorganisms, which performed specific, predictable tasks not normally executed by natural biological systems.
For example, these engineered organisms could be made to detect arsenic levels in drinking water; they could be formed into thin films on which visual patterns and images could be deposited, after the manner of a photographic film; they could produce hemoglobin that could be used to replace red blood cells in emergency transfusions. Or, just possibly, they could be fashioned into automated weight-control devices.
These new microbes were machines in more than a merely metaphorical sense. For one thing, they were built out of standardized parts, prefabricated genetic building blocks that could be reassembled in almost any desired order or combination. For another, these parts were to be put together according to a uniform or standard protocol for connecting one component to the next. In consequence, if the resulting combination of prefabricated, standardized biological parts worked at all, it worked predictably and reliably, just like a machine. That, anyway, was the idea.
To bring the idea of standardization into genomic engineeringâthe standardization of genetic parts, assembly methods, and combination procedures of the type advocated in the BioFab Manifestoâwould change the way we viewed, understood, and used biological systems. Originally, you pretty much had to take organisms as they came, with all the inherent design flaws and limitations, compromises and complications, that resulted from the random workings of evolution. Now we could actually preplan living systems, design them, construct them according to our
wishes, and expect them to operate as intendedâjust as if they were in fact machines,
appliances
.
These ideasâorganisms as machines, part by part construction, standardizationâhave their counterparts in the third industrial revolution, which is the one we normally think of when we use the term “industrial revolution.”

The third industrial revolution (1750â1850) was one of the great turning points in human history. Key elements of the transformation were the change from artisanal, custom-made, hand-tooled methods of producing material goods to machine-tooled, assembly-line, and standardized mass production systems. These changes allowed for unprecedented levels of income growth and wealth accumulation, sustained increases in agricultural production, human population growth, and enhanced well-being.
The third industrial revolution was a product of at least two separate technological developments. The first was the use of steam to power machinery, and this goes back to ancient times. It began over the period from 250
BCE
to 50
CE
with Ctesibius (pronounced
teh-sib'-ee-us)
, and then Hero (his real name) and his “aeolipile” device (its real name). Ctesibius, an inventor and engineer living in ancient Alexandria, was a maestro of air and water. He built an air-powered catapult, a water-powered pipe organ, and a water clock that worked by dripping water into a container at a constant rate: a float with a pointer attached to it marked the passage of time on a vertical scale. He also described a device for pumping water out of wells.
But it was Hero of Alexandria who invented one of the first steam engines. He attached two bent tubes to a hollow sphere, filled it with water, and heated it over a fire. The tubes then became steam nozzles and caused the sphere to spin (this was his aeolipile). Although the device did no practical work, it was nevertheless an apparatus that converted steam into motion, and was thus a true machineâa steam turbine. (Among other things, Hero also invented the world's first vending machine, a Rube Goldberg
apparatus in which a set amount of holy water would be dispensed after a coin was deposited through a slot at the top.)
Practical use of steam awaited Englishmen Thomas Savery in 1698 and Thomas Newcomen in 1712. Savery's steam engine had no piston or other moving parts and worked instead by generating steam inside a vessel and letting it condense by cooling, which induced a partial vacuum that drew water up through a tube.
Newcomen constructed a mechanical engine in which a giant arm was made to reciprocate back and forth by filling a chamber with steam at about ten times atmospheric pressure. This drove a piston up inside the chamber, raising one end of the arm until, at the top of the stroke, the pressure was released through a valve. The process was then repeated. These devices were typically used for pumping water out of mines and were widely employed and widely imitated. In 1775 James Watt, the Scottish inventor and mechanical engineer, attempted to replace water as the obligatory source of power for mills. This freed the mills from the need to be located near streams.
The other development that made the industrial revolution possible was standardization. Earlier, when individual craftsmen turned out their wares, each item was a product of the artisan's own personalized set of hand toolsâchisels, files, scrapers, planes, hammers, saws, and so onâall of them used according that individual's particular level of skill, working methods, and often idiosyncratic design of the finished product. The result was a vast range of made-to-order manufactured objects, each of which was mechanically incommensurable with all the others. Even the very nuts and bolts, the screws and nails, that held the object together differed sufficiently from one clock maker or gunsmith to the next that in many cases one artisan could not repair an artifact made by another. This system was not a model of optimization.
But on April 21, 1864, William Sellers, a toolmaker who was also president of the Franklin Institute in Philadelphia, read before that society a paper that would become one of the landmark documents of the machine age. Titled “On a Uniform System of Screw Threads,” it proposed a system in which every nut, bolt, and screw would be of uniform length and
diameter, and the threads would be angled at a precise and consistent 60-de-gree pitch. Such threads were easy to measure and manufacture (60 degrees being the angle between the sides of equilateral triangles), and they made for an exceptionally strong and secure fit. This uniformity of size and thread made for a system of fully interchangeable parts.
The Sellers system soon became a benchmark, then a national standard, and finally an international standard. It was a paradigm case of sensitive dependence on initial conditions, for his seemingly fussy, minor, even trifling innovation was the crucial design change that allowed the hand tool age to give way to the machine tool era characterized by interchangeable parts, reliable operation, and assembly-line mass production.
In twenty-first-century America, the William Sellers of biology was an MIT computer science whiz, artificial intelligence engineer, and information omnivore by the name of Tom Knight. He arrived at CSAIL (MIT's Computer Science and Artificial Intelligence Laboratory) as a fourteen-year-old high school student one summer in the 1960s and apprenticed himself to legendary artificial intelligence sage Marvin Minsky. Knight would go on to invent, coinvent, and play supporting roles in developing a number of the iconic structures of the computer age, including hardware for ARPANET, LISP machines, and Danny Hillis's Connection Machine.
Later, in the 1990s, Knight came across the work of Harold Morowitz, a Yale physicist and biologist. (A man of considerable dry wit, Morowitz was the author of several popular science books, including
The Thermodynamics of Pizza
and
Mayonnaise and the Origin of Life
.) Morowitz had a special interest in a category of bacteria called (seriously)
Mollicutes
, the most generally known of which were the
Mycoplasmas
, which were also the smallest and simplest known free-living, self-replicating life forms. They consisted of a countable number of atoms (about a billion). Morowitz thought, and so did Knight, that studying these ultra-simple organisms would reveal the underlying architecture of life.
“Morowitz's work laid out, in words I could understand as an engineer, an agenda that seemed so exciting that I had to go do it myself” Knight once explained. “Here's a class of organisms so simple that maybe we can understand everything there is to know about them.”
Being very much a hands-on, let's-find-out type of guy, Knight set up a biology lab inside of a little niche within CSAIL where he could do experiments on
Mesoplasma
, a species closely related to
Mycoplasma
. He quickly discovered that organisms, even small and simple ones, did not operate with the predictable crisp precision of computer electronics or well-written software. And there was nothing routine or standardized about the process of experimenting on them.
Every time he tried to make a DNA construct for an experiment, he said, “it was done in a different way, depending on what plasmids were available, what restriction sites were present in the fragments being assembled, what cell lines were available for transformation. The list goes on and on, and the more you know, the longer the list is. It drove me crazy, as it would any self-respecting engineer. The path forward was clearly to standardize the part definition and the assembly process.”
So, in essence, that is what he did. In a paper titled “Idempotent Vector Design for Standard Assembly of Biobricks” (2003), Knight proposed the idea of a standard biological part, which he called a biobrick, as well as a standard procedure for putting two or more of them together. Each biobrick would be a circular piece of double-stranded DNA (otherwise known as a plasmid) containing a smaller sequence of DNA base pairs of known structure and function. The smaller sequence was the biological part of interest, whereas the rest of the plasmid was a sort of backbone or framework. Each end of the smaller sequence (the “prefix” and “suffix”) consisted of a specific molecular structure that made for a sticky end that could join with the corresponding sticky end of any other biological part.
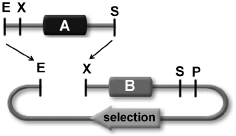
The standard procedure for assembling two biological parts was simple enough conceptually (
Figure 8.1
). Using the restriction enzymes and the cutting and joining techniques commonly employed in molecular biology, you would cut biological part
A
out of the plasmid that contained it, and slice open a space at the sticky-end site of the plasmid containing the second biological part,
B.
Then you would mix together part
A
and the sliced-open plasmid that contained part
B
, thus allowing the respective sticky ends of parts
A
and
B
to join up. This created an expanded plasmid containing the new biological part
AB.
That plasmid could then
be put into
E. coli
cells, which would express whatever it was that the new part coded for.