Life's Ratchet: How Molecular Machines Extract Order from Chaos (27 page)
Read Life's Ratchet: How Molecular Machines Extract Order from Chaos Online
Authors: Peter M. Hoffmann

FIGURE 6.3.
How an enzyme accommodates a transition state and therefore lowers the activation energy of a chemical reaction.
To illustrate the remarkable abilities of enzymes as catalysts, consider an enzyme called phosphoglucomutase. This enzyme converts an indigestible kind of sugar into a more palatable one. By speeding up the conversion process by a factor of one trillion, one phosphoglucomutase molecule can convert a hundred sugar molecules per second. Without this enzyme, it would take three hundred years to convert just one sugar molecule!
Enzymes are large molecules made of proteins—long chains of amino acids that fold into complicated shapes, as described in
Chapter 4
. Proteins are like a box of Legos: By changing the sequence of amino acids, almost any shape of protein can be constructed. Using the trial and error of evolution, myriads of enzymes have evolved, each custom-made to catalyze a particular reaction.
Molecular machines can be considered a special type of enzyme. As will be described later, they have evolved from enzymes. Enzymes and molecular machines share many attributes. In some sense, enzymes
are
machines: They bind, transform, and release molecules by virtue of their specific structure, but also as a result of being jostled by the molecular storm. A molecule does not know how it will fit into the binding pocket of the enzyme. Instead, the constant bombardment by water molecules (about every 10
−13
seconds) rapidly rotates and deforms the molecule. After a millisecond, by chance, the molecule is pounded into the right shape and orientation to form a complex with the enzyme. If you are amazed that the molecule is able to find the right orientation and shape, consider that in a millisecond, an average molecule undergoes ten billion collisions with water molecules.
Enzymes are amazing molecules, but they are not what we would call alive. First of all, they do not move. They deform a little bit when binding a substrate molecule, but this seems hardly sufficient to operate a molecular machine.
In the early days of biochemistry, biologists envisioned the cell as a pouch containing enzymes that catalyzed various reactions. The cell was thought of as nothing more than a tiny reaction vessel. It soon became clear, however, that cells would not survive long if enzymes were given free rein to make and break down molecules. Cells have to respond to their environment. Cells have to be able to make decisions.
Computers can make decisions because computer programs contain logical commands such as “IF . . . THEN. . . .” In a cell, logical decisions are implemented on a molecular scale. Part of this cellular software is, of course, DNA, which contains the blueprint for how to make different proteins and how to guide a body through its development. But DNA does not control the day-to-day operation of the cell. This is done by enzymes and molecular machines. So the real question is, how do enzymes compute?
This mystery was solved in the early 1960s by Jean-Pierre Changeux (b. 1936), a young graduate student in Jacques Monod’s laboratory at the Pasteur Institute in Paris. Studying an enzyme called L-threonine deaminase, which broke down its substrate, the amino acid threonine, he found that the enzyme’s activity was inhibited in the presence of the molecule isoleucine, another amino acid. Inhibition of enzymes had been seen before and was thought to occur when the enzyme was bound to a molecule other than its substrate. This process of an unwanted houseguest’s clogging up the binding pocket of an enzyme is called competitive inhibition. A familiar example is carbon monoxide poisoning: This gas will fit so strongly into the oxygen-binding pocket of hemoglobin (the oxygen-carrying proteins in our red blood cells) that it cannot be released again. In the competition between oxygen and carbon monoxide, carbon monoxide is the clear winner. Once the hemoglobin in a victim’s blood becomes gummed up with carbon monoxide, oxygen transport is shut down, and the hapless victim suffocates.
Changeux’s deaminase did not follow the model of competitive inhibition, however. When he tried to shut down the isoleucine binding site on the enzyme, the enzyme continued its enzymatic activity uninhibited. If the isoleucine binding site and the catalytic binding pocket had been one and the same—as in the case of hemoglobin—the enzyme’s ability to bind the threonine substrate should have been impeded. Changeux concluded that the enzyme had
two
binding pockets: one for threonine (the enzyme’s
substrate), and one for the isoleucine, which regulated the enzyme’s activity.
In his autobiographical paper “Allosteric Receptors: From Electric Organ to Cognition,” Changeux recalls that he first presented his startling findings at the Cold Spring Harbor Symposium on Quantitative Biology in 1961. Here, he told his fellow scientists that “the interaction between these two sites was indirect and transmitted by a conformational change of the protein molecule.” In other words, binding the isoleucine caused a large change in the shape of the enzyme, closing down the binding pocket for the substrate, and rendering the enzyme unable to process threonine molecules (
Figure 6.4
). The isoleucine acted as a control molecule—it was as if in a computer program, there were a line: “IF isoleucine is present THEN do not process threonine.”
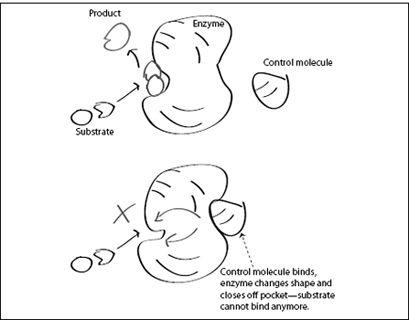
FIGURE 6.4.
Top: An allosteric enzyme combines two smaller molecules (its substrate) into one larger molecule by binding them to a suitable pocket in its structure (active site). A control molecule (right) approaches the enzyme. Bottom: The control molecule has attached itself to the enzyme’s control site. This has caused the enzyme to change shape (conformation) and close off the active site. The enzyme halts its catalytic activity as long as the control molecule remains bound.
Much enzymatic activity in living cells is controlled by the binding and release of such control molecules. Binding a control molecule changes an enzyme’s shape, with part of the enzyme moving relative to other parts. This change in shape can increase or decrease the enzyme’s catalyzing activity by opening or closing the pocket that binds the enzyme’s substrate. The control molecules can also completely shut down the enzyme. This ability of a control molecule to change the enzyme’s shape and activity was named
allostery
by Changeux’s thesis advisors, Jacques Monod and François Jacob.
Once you have allostery, you can implement many useful schemes to control chemical reactions in a cell. Imagine that an enzyme creates a product, which also serves as the enzyme’s control molecule. This would allow a product to regulate its own production. Such circular schemes are known as feedback loops. A familiar feedback loop is the feedback between an audio speaker and a guitar pickup—when a rock guitarist holds her guitar against the speaker, the sound from the speaker causes the guitar strings to vibrate. This is picked up by the pickup, amplified, and fed back to the speaker, which now makes an even louder sound, vibrating the guitar strings even more. The result is an increasingly loud shriek, only limited by the power of the amplifier. This kind of feedback is known as
positive feedback
—a feedback where the product (sound, in our example) enhances the production of more product. Positive feedback also exists in cells—some enzymes speed up production in the presence of product. The result is a rapid, explosive increase in product (until the reactants run out). This can be useful if the cell needs to produce a chemical very quickly in response to an external stimulus.
More common is the opposite case:
negative feedback
. An example is Changeux’s L-threonine deaminase. This enzyme is part of a number of enzymes that work together to make isoleucine, starting from threonine. But isoleucine was also the control molecule that inhibited the activity of L-threonine deaminase. Thus, the product inhibits its own production. As a result, the enzymes make just enough product until the product molecules shut down further production. This is a nice way to control the maximum amount of a product molecule in a cell.
There are more complicated schemes that involve vast networks of interacting enzymes. The product of one enzyme may act as the control
molecule for another enzyme, either enhancing or inhibiting its activity. The product of this second enzyme may again control the first enzyme, forming a two-enzyme feedback loop as shown in
Figure 6.5
, or the product may influence a third enzyme, which influences a fourth, and so on. Complicated schemes of feedback loops and mutual enhancement or inhibition provide the computing power that makes living cells seem intelligent.
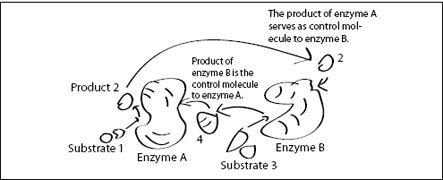
FIGURE 6.5.
A molecular feedback loop. The product 2 of enzyme A serves as a control molecule for enzyme B, while the product 4 of enzyme B serves as a control molecule for enzyme A. Depending on the control molecule’s influence on the enzymes (inhibition or enhancement), several programs can be implemented this way. In addition, substrates 1 and 3 may be the products of other enzymes, which themselves are controlled by control molecules. This way, more and more complex molecular programs can be constructed.
Allosteric enzymes bring us closer to our understanding of molecular machines. A large change in the shape of an enzyme, when it binds to a substrate or control molecule, can be seen as a type of motion. We could imagine, for example, that this change in shape creates forward motion when we place an allosteric enzyme on a molecular track: Each time the enzyme changes shape, it pushes against the track and propels itself forward (
Figure 6.6
). Examples of molecular tracks include fibers and filaments, typically made of long strands of proteins. One example is actin, which we already encountered in
Chapter 4
. Another example is DNA— and indeed there are specialized machines that move along DNA like locomotives
on a train track. We will meet some of these machines in the next chapter.
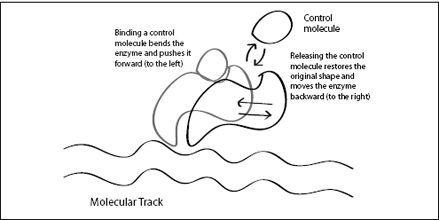
FIGURE 6.6.
An allosteric enzyme bound to a molecular track. As the control molecule binds, the enzyme changes shape and pushes the enzyme forward along the track. However, the unbinding of the control molecule pushes the enzyme back to where it came from. To move forward, it would need to detach temporarily.
A molecular machine, however, is not simply an allosteric enzyme bound to a track. The problem with this idea is that an allosteric enzyme exists in exactly two states: It is in shape A if the control molecule is attached, and shape B when the control molecule is released. The enzyme may push itself forward when binding a control molecule, but would revert to its old shape and move backward once the control molecule is released. Just as with our ratchet in
Chapter 5
, the molecule would dither back and forth, but make no headway. To move forward, the machine needs to loosen its grip on the track during the back step and only attach during the forward step. But if the machine loses its grip on the track, the molecular storm will sweep it away.