Out of Eden: The Peopling of the World (9 page)
Read Out of Eden: The Peopling of the World Online
Authors: Stephen Oppenheimer

Figure 0.3
Real maternal gene tree of 52 randomly selected individual people from around the world. Note the age of Mitochondrial Eve. Branch dating by author based on complete sequence data; the chimp–human coalescent date arises from analysis, i.e. not assumed from fossil evidence – see note
22
in
Chapter 1
.
Y chromosome: the Adam gene
Analogous to the maternally transmitted mtDNA residing outside our cell nuclei, there is a set of genes packaged within the nucleus that is only passed down the male line. This is the Y chromosome, the defining chromosome for maleness. With the exception of a small segment, the unpaired Y chromosome plays no part in the promiscuous exchange of DNA indulged in by other chromosomes. This means that, like mtDNA, the non-recombining part of the Y chromosome remains uncorrupted with each generation, and can be traced back in an unbroken line to our original male ancestor.
Y chromosomes have been used for reconstructing trees for less time than mtDNA has, and there are more problems in estimating time depth. When these are solved, the NRY method may have a much greater power of time and geographical resolution than mtDNA, for both the recent and the distant past. This is simply because the NRY is much larger than mtDNA and consequently has potential for more variation.
Yet Y chromosomes have already helped to chart a genetic trail parallel to the mtDNA trail. At the major geographical branch points they support the story told by mtDNA: they point to a shared ancestor in Africa for all modern humans, and a more recent ancestor in Asia for all non-Africans. In addition, because men’s behaviour differs in certain key ways from women’s, the story told by the Adam genes adds interesting detail. One difference is that men have more variation in the number of their offspring than women: a few men father considerably more children than the rest. Women, in contrast, tend to be more even and ‘equal’ in the number of children they have. The main effect of this is that most
male lines become extinct more rapidly than female lines, leaving a few numerically dominant male genetic lines.
Another difference is in movement. It has often been argued that because women have usually travelled to their husband’s village, their genes are inevitably more mobile. Paradoxically, while this may be true within one cultural region, it results in rapid mixing and dispersal of mtDNA only within that cultural region. For travel
between
regions, or long-distance intercontinental migrations, by sea for example, the burden of caring for children would have limited female mobility. Predatory raiding groups would also have been more commonly male-dominated, resulting in increased long-distance mobility in the Y chromosome.
A final point on the methods of genetic tracking of migrations: it is important to distinguish this new approach to tracing the history of molecules on a DNA tree, known as phylogeography (literally ‘tree-geography’), from the mathematical study of the history of whole human populations, which has been used for decades and is known as classical population genetics. The two disciplines are based on the same Mendelian biological principles, but have quite different aims and assumptions, and the difference is the source of much misunderstanding and controversy. The simplest way of explaining it is that phylogeography studies the prehistory of individual DNA molecules, while population genetics studies the prehistory of populations. Put another way, each human population contains multiple versions of any particular part of our genome, each with its own history and different origin. Although these two approaches to human prehistory cannot represent exactly the same thing, their shared aim is to trace human migrations. Tracing the individual molecules we carry is just much easier than trying to follow whole groups.
Naming gene lines
In this book I refer interchangeably to maternal or paternal clans, gene lines, lineages, genetic groups/branches, and even haplo-groups.
All these terms mean much the same thing: members of a large group of genetic types sharing a common ancestor (usually through their mtDNA or Y chromosome). The size of the group is to a certain extent arbitrary and depends on how far back the base of the branch is on the genetic tree. One thing that quickly becomes apparent from study of genetic trees is the lack of uniformity in the nomenclature of these branches. For the Y chromosome in particular, each new scientific paper proposes a new system of scientific nomenclature based on different ‘in-house’ genetic markers used by different research laboratories. This can make comparison between different studies tedious and repeatedly tests the limits of the reader’s memory. The underlying tree, however, is more or less the same from lab to lab. Recently, a consensus Y nomenclature was published, with letters from A to R describing the main genetic branches in the tree.
37
I use this nomenclature as far as possible in my referencing and figures. The trouble is that even these eighteen letters and their location on the tree are difficult to hold in the mind – at least in mine. The fact that they are just letters makes it worse.
Luckily, our memory is often aided by context and association. For this reason, and this reason alone, I have introduced names for the major branches and for other branches I refer to frequently. Some of the names are regional, such as Ho for one Chinese/Southeast Asian Y-branch (after the Southeast Asian explorer Admiral Cheng Ho and also for Ho Chi Minh, the revered Vietnamese nationalist hero). Others are biblical, like the out-of-Africa Y-line founder Adam and his three descendent lines, Cain, Abel, and Seth (see
Appendices 1
and
2
). There is no intention with these names to infer any deeper meaning – they are simply aides-mémoires.
The mtDNA picture is slightly easier. Many of the different labs agreed at an early stage to try to use a single nomenclature. (Perhaps there was less testosterone involved in the process!) For instance, there are two agreed non-African daughter lines, ‘N’ and ‘M’ from the single out-of-Africa line L3. I have called them Nasreen, in keeping with a southern Arabian origin, and Manju, to be consistent with an Indian subcontinental origin.
O
UT OF
A
FRICA
O
NE OF THE MOST ENDURING
media images of popular genetics in the 1980s was a cover of
Newsweek
showing sophisticated and attractive nude portraits of a black Adam and Eve sharing the apple, with the snake looking on approvingly between (see
Plate 5
). This cover sold record numbers of the magazine. But in spite of the media hype,
Newsweek
was reporting a major advance. There were two stunning insights portrayed in the picture and described in the lead article. The first was some new genetic evidence published in 1987, using genes that could be passed down only through our mothers. This work, by American geneticist Rebecca Cann and colleagues, resolved an old argument about the birthplace of modern humans. The new evidence said that we, ‘the modern human family’, had originated as a single genetic line in Africa within the last 200,000 years, and not as multiple separate evolutionary events in different parts of the world. This single line, which leads back eventually to the ancestor we share with the Neanderthals, gave rise to the half-dozen major maternal clans (or branches) that are, even today, clearly of African origin.
1
The second reason for using a biblical allegory on the
Newsweek
cover was that this new genetic approach used only maternally
transmitted mitochondrial DNA. Ten years later, a small group of geneticists would use this newly discovered method to identify a single twig from those dozen or more ancestral African maternal genetic branches as forming the sole founding line for the rest of the world.
2
In other words, there was a single common ancestor or ‘Mitochondrial Eve’ for all African female lines and then, much later, came a subsidiary ‘Out-of-Africa Eve’ line whose genetic daughters peopled the rest of the world. It proved to be an extraordinary discovery.
The label ‘Eve’ or ‘First Lady’, so celebrated by the media at the time, was not the exact truth as geneticists saw it. Using mitochondrial DNA, they had identified a root female genetic line for only a tiny part of our genetic heritage. Mitochondrial Eve was not a sole individual ‘First Mother’ as often implied by the media. Tracing a maternal genetic marker back to a shared ancestral type does not mean that
all
our genes literally derive from one woman. In each of our cell nuclei we have tens of thousands of genes, each with their own history, that can be mapped. Any one of these genes could be used as a marker system to trace back to a common ancestor.
3
Because much of our DNA mixes around at each generation, the gene trees would not necessarily all go back to the same ancestor. In fact, the genetic heritage of modern humans may be derived from a core of 2,000–10,000 Africans who lived around 190,000 years ago.
Although the reality of parallel ‘gene trees’ rather than ‘people trees’ may seem to take the romance out of the Eve tale, it does not diminish the exceptional power of such genetic tracing to tell the grand story of human wanderings over the past 200,000 years. The ability to tell the same story using a number of different genes confirms and enriches the tale. Male gene lines, for instance, tend to show a rather more adventurous intercontinental spread than do female ones. In South Africa, for example, a self-proclaimed ‘lost tribe of Israel’ was recently identified by geneticists as having Jewish ancestors, but only through the male line.
4
Objections from multiregionalists and geneticists
The bold and clear trail of spreading humankind drawn by the mitochondrial markers was bound to raise objections. The multiregionalists certainly dissented. These were mainly palaeoanthropologists who still believed that the different world ‘races’ each broke out of pre-modern ancestral forms – from various parts of the world such as ‘Java Man’, ‘Peking Man’, and the Neanderthals. A major problem here is that, in many ways, the regionally defined modern human peoples resemble one another more than they do their supposed ancestors. To cope with this observation, some multiregionalists now argue that different regional ‘human species’ subsequently mixed and exchanged genetic material with one another to form the complex modern races we have today (
Figure 1.1
).
Yet the genetic evidence fails to show such fine inter-regional mixing. The geographical distributions of the branches and twigs of the modern out-of-Africa mitochondrial DNA and Y-chromosome trees are regionally very specific. In a last stand by the multiregionalists, the ‘inter-regional mixing compromise’ has been extended further to encompass the out-of-Africa hypothesis by suggesting that regional archaic human populations, such as Neanderthals, occasionally interbred with incoming anatomically modern – that is, ‘like us’ – Cro-Magnons. (‘Anatomically Modern Humans’ are members of the species
Homo sapiens
(
sensu stricto
), possessing a combination of skeletal features that collectively distinguish ourselves and our immediate ancestors in Africa within the past 200,000 years from extinct archaic versions of
Homo sapiens
(
sensu lato
), including our relation
Homo helmei
. The main ‘anatomically modern’ characteristics are a high round cranium, a pronounced chin, small face and jaws, and brow ridges.) Inter-regional mixing of old-timers and newcomers is in effect a soft compromise of the out-of-Africa hypothesis, which would allow some leftover genes from archaic peoples to be present in the modern gene pool. A child’s
skeleton in Portugal, showing short robust limbs and dated to 24,500 years ago – after Neanderthals died out – has been cited as evidence of such hybridization. Yet, again there is no evidence among the tens of thousands of non-Africans who have had their mtDNA and Y chromosomes studied for even a minimal degree of this kind of mixing.
5
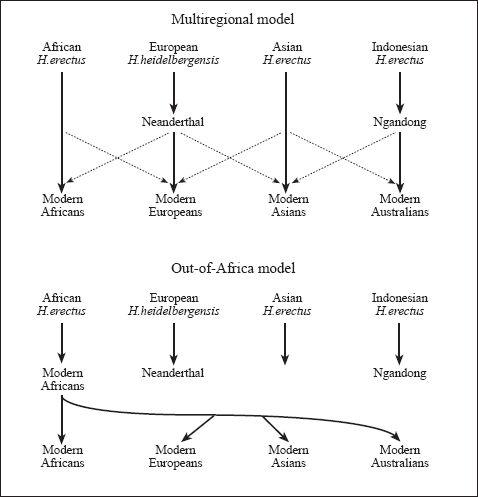
Figure 1.1
Comparison of multiregional and out-of-Africa models. In the out-of-Africa model ‘Anatomically Modern’ Africans recently completely replaced all other humans worldwide; in the multiregional hypothesis a variable degree of genetic and physical inheritance remains today from older regional species of humans.
To be fair, that is not the end of the story. While there is no evidence in the ‘Adam’ and ‘Eve’ lines of any such modern/archaic admixture, these lines hold but a tiny fraction of our huge genomic library of DNA. Because only one version each of mtDNA and the Y chromosome (NRY) are transmitted at each generation, rather than two, Neanderthal-specific mtDNA lines may on a very few occasions have admixed into Cro-Magnon populations and then become extinct. But there could still be evidence of miscegenation lurking among the rest of the vast nuclear genome. Because such nuclear DNA shuffles and splices at each generation, it is harder to build a tree for each of the numerous gene lines and be sure what was brought in by modern humans and what derives from regional archaic peoples.
6
So it could just be that, as some people claim, the beetle-browed appearance of some rugby internationals and soccer hooligans eventually turns out to be a Neanderthal throwback, rather than the more likely event (in my view) of normal variation in modern humans.