Whole (19 page)
Authors: T. Colin Campbell

Despite decades of disappointment, most of us still believe in the Big Promise of modern medicine: a world free from disease and early death, a paradise in which we no longer have to fear scourges like cancer, heart disease, diabetes, and so on.
To understand why we believe this, you need only look at the remarkable advances of twentieth-century medical science. In 1900, medicine could not reliably cure infection, transplant organs, keep people alive on respirators, replace failing kidneys with dialysis, or look deeply into our bodies with MRI and CT scans. The list of recent medical advances leads us to believe that our progress has been staggering. Why wouldn’t we assume that future breakthroughs will be even more remarkable? As computers and other technologies advance, it just makes sense that someday soon, all these discoveries and inventions will save us from both our folly and most, if not all, of the diseases that still plague humankind.
The medical establishment has fanned the flames and basked in the glow of our love affair with scientific progress. After all, our collective
faith in the Big Promise has funded the War on Cancer, among many others. And popular culture has enshrined the image of the selfless, heroic researcher hot on the trail of the “cure” for cancer.
Trouble is, the medical establishment hasn’t had any real wins in a long time. Technology has advanced at a breakneck pace, but technologies that actually improve health outcomes have been hard to find. While death rates in developed countries plummeted in the early part of the twentieth century largely due to an understanding of hygiene,
1
none of the ultra-expensive high-tech advances of the past fifty years have made a dent in overall rates of death and disease in first-world countries. And while medicine is now much better equipped to save someone’s life after an acute event like a car crash or a sudden heart attack than it was fifty years ago, we’re really no better at preventing chronic degenerative diseases like heart disease and cancer, often called “diseases of affluence,” than we were in the 1950s.
Yet we still look for the next medical knight on a white horse to ride to our rescue: the pill, the vaccine, the technology, the intervention that will disease-proof us and save us, not just from the diseases themselves, but from the pervasive fear of diseases that seem to strike randomly in our midst.
It’s the (apparent) randomness that scares us the most. I remember the fallout when Jim Fixx, author of the 1977 bestseller
The Complete Book of Running,
died of a heart attack at the age of fifty-two. The media reported his death with an air of ironic fatalism, as proof that death would find us no matter how fervently we pursued a healthy lifestyle.
What we really want from science is an end to randomness. We want to know why diseases strike some people and not others. We want to know how to protect ourselves against the scourges that have our names on them. We want, in short, to banish unpredictability.
In a reductionist universe, you’ll recall, unpredictability is not allowed. In a universe that is simply a mechanical expression of physical laws, everything is theoretically knowable. If we can’t predict in advance exactly who will get pancreatic cancer or heart disease, it’s simply because we haven’t collected enough data yet. We don’t yet have tools sensitive or powerful enough to lay bare the apparent mystery. But no fear—they’re coming! In fact, they’re just about here! The problem is, they’ve been “just about here” for the last forty years or so.
In recent years, one discipline has gained prominence over the rest as the one that will solve all our health problems and tell us all those things we don’t yet know. I’m speaking, of course, of the genetics revolution that began in the early 1950s and has been gathering steam (and money) ever since. You could argue that we are now living in the Age of Genetics. The mapping of the human genome and individual gene sequencing are the cutting edge of medical technology. DNA is the master code, right? Our entire biography and destiny, mapped out in a fantastically long and complicated blueprint. All the secrets of our development and our nature are contained in that DNA double helix: our physical appearance and function, our personality, our predisposition to various diseases. As computing power and speed increase, we continue to unravel these secrets. Soon, as a March 7, 2012
New York Times
article trumpeted, the cost of individual gene sequencing will be as modest as that of a simple blood test, with “enormous consequences for human longevity.”
2
The scientists at the Silicon Valley start-ups behind this push for fast, affordable sequencing operate from the assumption that the limiting factor in improving human health has been a lack of data. Typical of this faith is the statement of Larry Smarr, director of the California Institute of Telecommunications and Information Technology and a member of the scientific advisory board for Complete Genomics (one of Silicon Valley’s gene sequencing pioneers): “For all of human history, humans have not had the readout of the software that makes them alive. Once you make the transition from a data poor to data rich environment, everything changes.”
3
These genetic crusaders view themselves as pioneers in a new age of enlightenment—specifically, reductionist enlightenment. Genes, in the genetic crusaders’ view, are simply human software. Just as a good programmer can read code and predict exactly what the program will do, eventually we’ll be able to look at genes and predict exactly what diseases we’ll develop, perhaps even what emotions we’ll experience from moment to moment.
The problem is, we can’t. Genes tell us what may happen, but not if or how. The increasing fascination with and funding of genetic technology is simply another medical dead end, another reductionist rabbit hole that will lead us no further toward preventing and reversing chronic illness.
As with nutrition, the discipline of genetics is unimaginably complex. This complexity has not filtered down to the public. Most of the population tends to think of genes as relatively fixed entities that cause us to look and function and behave in particular ways. The truth is far more interesting.
When I was on the farm, my brothers, Jack and Ron, and I each had a “self-propelled combine”—a big machine that harvested grain as we drove through the field (our way of helping our father earn money for our college education). In those days, combines were about as mechanically complex as any other machine on the market. I’ve forgotten how many belts and pulleys there were on my machine, but I remember well the 103 fittings that I had to fill with grease at the start of each and every day. For me it was an engineering marvel of ordered complexity. But these machines were only the beginning of the engineering marvels yet to come: ever-larger airplanes, massive ocean liners, talking radios in color (i.e., TVs), satellites and space stations, communication devices and systems, really fancy laboratory equipment, and now computers everywhere. Marvelous machines, marvelous minds! But as impressive as these engineering and technical feats may be in their complexity and order, they pale into insignificance when compared with the microcosmic universes of molecular genetics.
As you may remember from high school biology, DNA is a long thread composed of two parallel strands that are gently twisted together into a double helix shape. Alternating sugar and phosphate molecules link to form the backbones of these adjacent strands (seen as ribbons in
Figure 8-1
).
Strung along these strands are four precisely arranged, or sequenced, nitrogen-containing bases, each of which is anchored to a deoxyribose unit of the strand. They are named adenine (A), thymine (T), guanine (G), and cytosine (C), and they project perpendicularly from each strand in a way that faces partner bases on the adjacent strand, thus facing inward
and holding the strands together. The facing As and Ts of each strand have a chemical affinity for each other, thus forming
base pairs
; Gs and Cs form similar pairs.
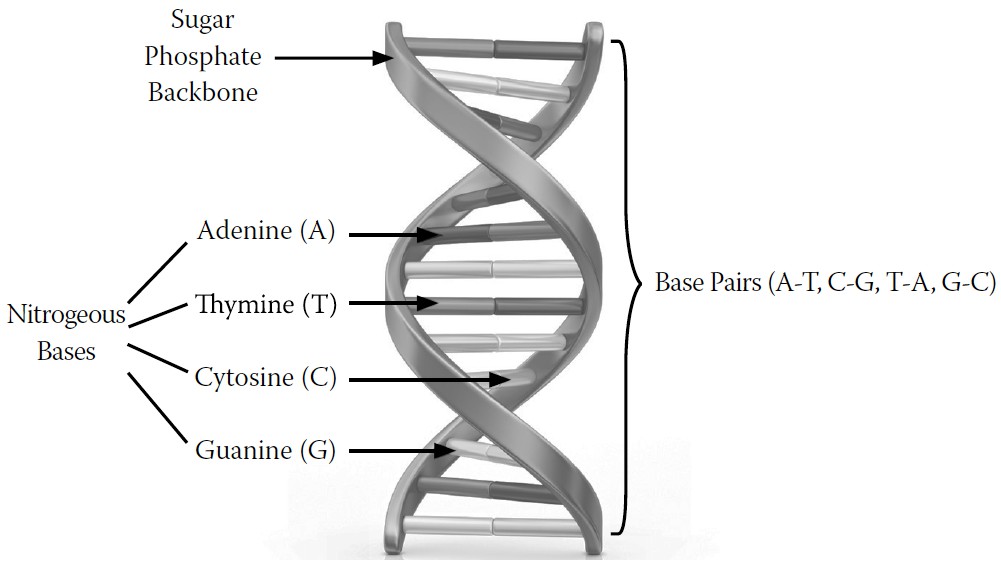
FIGURE 8-1.
A DNA molecule
The DNA molecule is unimaginably long and harbors these four bases in a sequence that is unique for each and every person who ever lived on the planet. Because these bases act like letters of an alphabet that create words, they have the capacity to create an enormous body of information.
4
This unique DNA chain is clipped and packed into twenty-three pairs of chromosomes located within the nucleus of each of the 100 trillion cells in our bodies (which, individually, are small enough to sit comfortably on the tip of a pin). Our cells use DNA as a blueprint for doing their work. The bases on the twenty-three chromosome pairs (about three billion bases, in total) are grouped into aggregates (around 25,000 of them) called
genes.
And each of these genes, which may contain as few as 100 bases and up to as many as several million, ultimately directs the formation of a unique protein.
However, these genes do not translate into a protein directly. Instead, they do so through the intermediate formation of ribonucleic acid (RNA) (
Figure 8-2
), a similar strand of bases that mirrors a DNA strand.
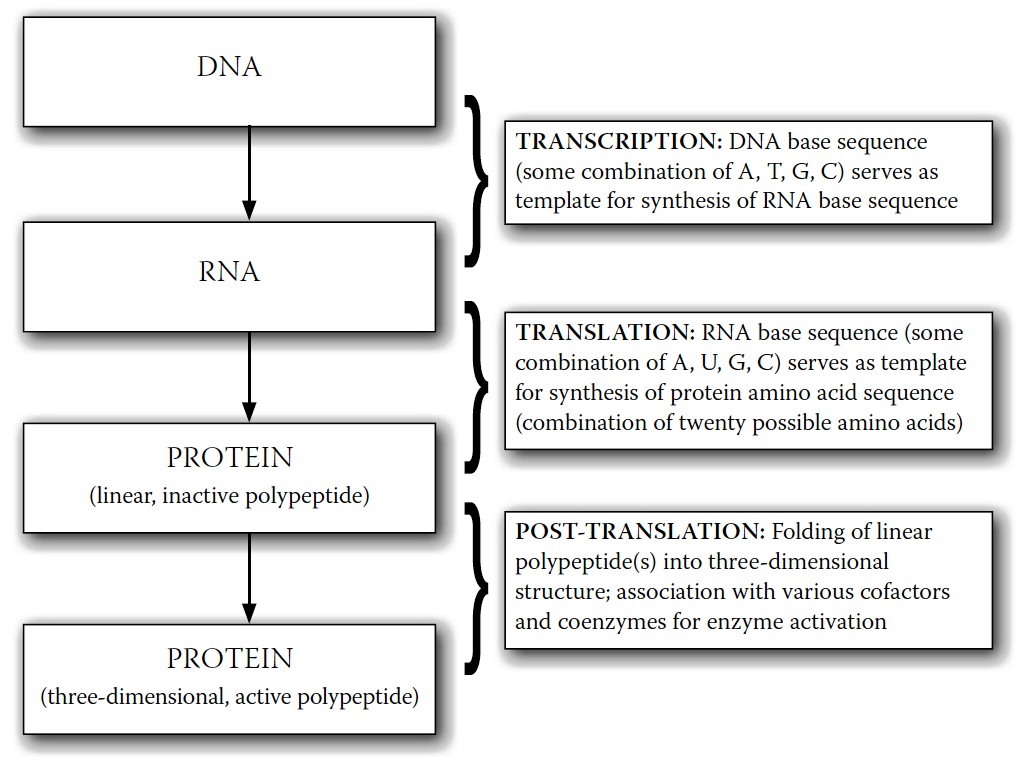
FIGURE 8-2.
The process of DNA expression into active proteins (e.g., enzymes)
The RNA base sequence serves in turn as a code for the selection of amino acids (about twenty amino acids are used in human protein production, each possessing a unique chemical structure) which, when combined into a long strand, form proteins. The bases on the RNA chains don’t code for these amino acids on a one-to-one basis, however. Instead, triplet sets of bases are used, each specifying one or more amino acids. With four bases, it is possible to create sixty-four different triplet combinations or
codons
(some amino acids can be specified by more than one triplet codon).
In the early days of genetic research, scientists believed in a “one gene/ one protein” hypothesis, in which each gene was responsible for expressing a single protein. If there were 25,000 genes, then that meant there were 25,000 proteins. However, recent work in the field makes it clear that this hypothesis is too simple. For instance, more than one gene can share in the making of a single protein, because some proteins are made
up of more than one strand of amino acids, and each of those amino acid strands is produced by a separate gene. The number of possible proteins and their combinations is impossible to estimate. The complexity at this point is far beyond comprehension by the human mind.
And here’s another puzzle. Despite the fact that each of our cells contains the exact same genetic master template as every other cell in our bodies, these cells can do very different things. A liver cell is very different from a nerve cell or a cell on the inner surface of the intestine, both in form and in function. Their structural and functional differences depend solely on which segments of DNA bases are selected for expression within each cell. The act of selecting which bases to use among the three billion bases is an awesome display of nature at work.
To recap: relatively short segments of the DNA base sequence, called genes, are transcribed into comparable RNA sequences, which translate, in turn, into sequences of amino acids that are used to make proteins. These proteins then provide the structure and function of cells, acting as enzymes, hormones, and structural units. It is through the activity of these proteins that DNA manifests its destiny.
That manifestation of destiny—the expression of genes, how they do what they do—operates through a series of enormously complex but very orderly processes. To investigate and understand these processes, researchers like to simplify them by thinking of seemingly discrete stages or events operating one after another, like dominos falling in a row. This simplification is helpful because it allows the details of each stage to be more easily investigated and visualized, but it is not entirely reliable. In reality, these stages or events are highly interconnected and communicative, a virtually seamless and extensively integrated stream of activities.